Article #:
0217-025
Antigen/Product:
Collagen type I, cleavage site
Synonym(s):
Col1-¾C
Also recognized:
Alpha-2 collagen type I (¾ fragment, C-terminal cleavage site, Gly775–Leu776)
Uni-/SwissProt #:
Clone / Name:
C o l 1 - ¾ C
Host Species:
rabbit
Mono-/polyclonal:
polyclonal (pAB) | epitope selective
Isotype:
n/a
Purity:
affinity purified on the antigen
Quantity (mass):
25 µg
Quantity (vol.):
100 µl
Price (net):
please enquire
Cross-reactivity:
human, mouse, rat, guinea pig, dog, cat, donkey, pig, cow, sheep, chicken, others not tested
Applications:
WB - Western blot / immunoblot, IF - immunofluorescence
Sugg. dilutions:
IF: 0.5 µg/ml - 2 µg/ml
Fixatives tested:
formaldehyde
Remarks:
Background Information
The proteolysis of collagens plays an important role in numerous physiological and pathological situations such as morphogenesis, wound healing, arthritis, arteriosclerosis, and tumor metastasis. Triple helical type I collagens are made up of two α 1 (I) and one α 2 (I) chains, and are found in skin, tendon, ligament and interstitial tissues. Due to their fibrillary structure native collagens are resistant to most proteases. They are substrates however for certain matrix metalloproteinases (MMPs), which constitute a family of zinc-dependent enzymes catalyzing the degradation of extracellular matrix components [1,2]. Initial MMP-8 dependent cleavage of collagen into the characteristic ¾ and ¼ fragments has been shown to enable MMP-9 diffusion along the protein helix, with preferential binding to the collagen ¾ fragment tail. Finally, untwisting of the helix end results in the local denaturation of the triple helical structure [3].
- [1] Song F., Wisithphrom K., Zhou J. & Windsor L. J. (2006). Matrix Metalloproteinase Dependent and Independent Collagen Degradation. Frontiers in Bioscience, 11:3100-20.
- [2] Bertini I., Fragai M., Luchinat C., Melikian M., Toccafondi M., Lauer Ja. L. & Fields G. B. (2012). The Structural Basis for Matrix Metalloproteinase 1 Catalyzed Collagenolysis. J. Am. Chem. Soc. 134(4): 2100–2110.
- [3] Rosenblum G., Van den Steen P. E., Cohen S. R., Bitler A., Brand D. D., Opdenakker G. & Sagi I. (2010). Direct Visualization of Protease Action on Collagen Triple Helical Structure. PLoS ONE 5(6): e11043.
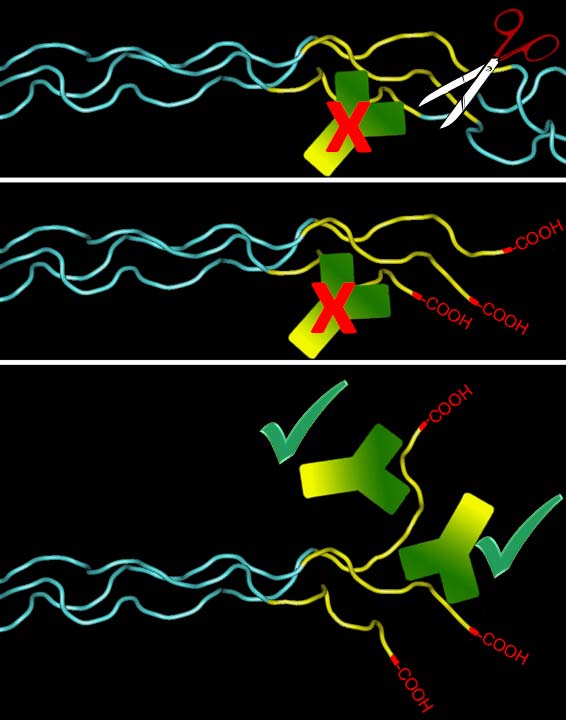
Model depicting antibody
detection of Col1 ¾C. MMPs cleaving the α chains, create free COOH
groups at the C-terminal end of the ¾ fragment, which gets untwisted and
exposes the antibody epitope. The carboxyl group proper is not part of
this epitope. However, there is also a companion antibody available
(IG-1266) that requires the free carboxyl group for binding (please
enquire).
Form:
sterile filtered liquid, with sodium azide, stabilized with carrier
Immunization:
synthetic peptide
Epitope:
C-terminal end of the N-terminal three quarter collagen fragment (Col1 ¾), which results from MT1-MMP, MMP-1, MMP-2, or MMP-8 dependent cleavage of the α 1 (I) and α 2 (I) chains at the G775–I776 & G775–L776 bonds, respectively
Storage:
-20°C
Shipping:
RT - ambient temperature
Availability:
in stock
Data sheet:
Publications referring to this product:
- Wang, Q., Ji, C., Ali, A., Ding, I., Wang, Y. & McCulloch, C.A. (2024) TRPV4 Mediates IL-1-Induced Ca2+ Signaling, ERK Activation and MMP Expression. The FASEB Journal, 38, e23731. https://doi.org/10.1096/fj.202400031R.
- Sabeh, F., Li, X.-Y., Olson, A.W., Botvinick, E., Kurup, A., Gimenez, L.E., Cho, J.-S. & Weiss, S.J. (2024) Mmp14-Dependent Remodeling of the Pericellular-Dermal Collagen Interface Governs Fibroblast Survival. The Journal of Cell Biology, 223, e202312091. https://doi.org/10.1083/jcb.202312091.
- Remy, D., Antoine-Bally, S., de Toqueville, S., Jolly, C., Macé, A.-S., Champenois, G., Nemati, F., Brito, I., Raynal, V., Priya, A., Berlioz, A., Dahmani, A., Nicolas, A., Meseure, D., Marangoni, E. & Chavrier, P. (2024) TFEB Triggers a Matrix Degradation and Invasion Program in Triple-Negative Breast Cancer Cells upon mTORC1 Repression. Developmental Cell, S1534-5807(24)00727–5. https://doi.org/10.1016/j.devcel.2024.12.005.
- Ranamukhaarachchi, S.K., Walker, A., Tang, M.-H., Leineweber, W.D., Lam, S., Rappel, W.-J. & Fraley, S.I. (2024) Global versus Local Matrix Remodeling Drives Rotational versus Invasive Collective Migration of Epithelial Cells. Developmental Cell, S1534-5807(24)00721–4. https://doi.org/10.1016/j.devcel.2024.11.021.
- Driscoll, M.K., Welf, E.S., Weems, A., Sapoznik, E., Zhou, F., Murali, V.S., García-Arcos, J.M., Roh-Johnson, M., Piel, M., Dean, K.M., Fiolka, R. & Danuser, G. (2024) Proteolysis-Free Amoeboid Migration of Melanoma Cells through Crowded Environments via Bleb-Driven Worrying. Developmental Cell, 59, 2414-2428.e8. https://doi.org/10.1016/j.devcel.2024.05.024.
- Coelho, N.M., Riahi, P., Wang, Y., Ali, A., Norouzi, M., Kotlyar, M., Jurisica, I. & McCulloch, C.A. (2024) The Major Vault Protein Integrates Adhesion-Driven Signals to Regulate Collagen Remodeling. Cellular Signalling, 124, 111447. https://doi.org/10.1016/j.cellsig.2024.111447.
- Remy, D., Mace´, A.-S., Chavrier, P. & Monteiro, P. (2023) Invadopodia Methods: Detection of Invadopodia Formation and Activity in Cancer Cells Using Reconstituted 2D and 3D Collagen-Based Matrices. Cell Migration in Three Dimensions, 225–246. https://doi.org/10.1007/978-1-0716-2887-4.
- Monteiro, P., Remy, D., Lemerle, E., Routet, F., Macé, A.-S., Guedj, C., Ladoux, B., Vassilopoulos, S., Lamaze, C. & Chavrier, P. (2023) A Mechanosensitive Caveolae–Invadosome Interplay Drives Matrix Remodelling for Cancer Cell Invasion. Nature Cell Biology, 25, 1787–1803. https://doi.org/10.1038/s41556-023-01272-z.
- Kang, L., Ikeda, S.-I., Yang, Y., Jeong, H., Chen, J., Zhang, Y., Negishi, K., Tsubota, K. & Kurihara, T. (2023) Establishment of a Novel ER-Stress Induced Myopia Model in Mice. Eye and Vision (London, England), 10, 44. https://doi.org/10.1186/s40662-023-00361-2.
- Ham, S.M., Song, M.J., Yoon, H.-S., Lee, D.H., Chung, J.H. & Lee, S.-T. (2023) SPARC Is Highly Expressed in Young Skin and Promotes Extracellular Matrix Integrity in Fibroblasts via the TGF-β Signaling Pathway. International Journal of Molecular Sciences, 24, 12179. https://doi.org/10.3390/ijms241512179.
- Beunk, L., Wen, N., van Helvert, S., Bekker, B., Ran, L., Kang, R., Paulat, T., Syga, S., Deutsch, A., Friedl, P. & Wolf, K. (2023) Cell Jamming in a Collagen-Based Interface Assay Is Tuned by Collagen Density and Proteolysis. Journal of Cell Science, 136, jcs260207. https://doi.org/10.1242/jcs.260207.
- Su, H., Yang, F., Fu, R., Trinh, B., Sun, N., Liu, J., Kumar, A., Baglieri, J., Siruno, J., Le, M., Li, Y., Dozier, S., Nair, A., Filliol, A., Sinchai, N., Rosenthal, S.B., Santini, J., Metallo, C.M., Molina, A., Schwabe, R.F., Lowy, A.M., Brenner, D., Sun, B. & Karin, M. (2022) Collagenolysis-Dependent DDR1 Signalling Dictates Pancreatic Cancer Outcome. Nature, 610, 366–372. https://doi.org/10.1038/s41586-022-05169-z.
- Ostrowska-Podhorodecka, Z., Ding, I., Abbasi, S., Norouzi, M., Ali, A., Arora, P.D., Wong, T.H.F., Magalhaes, M. & McCulloch, C.A. (2022) Vimentin Regulates Collagen Remodeling Through Interaction with Myosin 10. Qeios. https://doi.org/10.32388/OP0XRM.
- Oprescu, A., Michel, D., Antkowiak, A., Vega, E., Viaud, J., Courtneidge, S.A., Eckly, A., de la Salle, H., Chicanne, G., Léon, C., Payrastre, B. & Gaits-Iacovoni, F. (2022) Megakaryocytes Form Linear Podosomes Devoid of Digestive Properties to Remodel Medullar Matrix. Scientific Reports, 12, 6255. https://doi.org/10.1038/s41598-022-10215-x.
- Nader, G.P. de F., Agüera-Gonzalez, S., Routet, F., Gratia, M., Maurin, M., Cancila, V., Cadart, C., Palamidessi, A., Ramos, R.N., San Roman, M., Gentili, M., Yamada, A., Williart, A., Lodillinsky, C., Lagoutte, E., Villard, C., Viovy, J.-L., Tripodo, C., Galon, J., Scita, G., Manel, N., Chavrier, P. & Piel, M. (2021) Compromised Nuclear Envelope Integrity Drives TREX1-Dependent DNA Damage and Tumor Cell Invasion. Cell, 184, 5230-5246.e22. https://doi.org/10.1016/j.cell.2021.08.035.
- Colombero, C., Remy, D., Antoine‐Bally, S., Macé, A., Monteiro, P., ElKhatib, N., Fournier, M., Dahmani, A., Montaudon, E., Montagnac, G., Marangoni, E. & Chavrier, P. (2021) mTOR Repression in Response to Amino Acid Starvation Promotes ECM Degradation Through MT1‐MMP Endocytosis Arrest. Advanced Science, 8, 2101614. https://doi.org/10.1002/advs.202101614.
- Cho, Y., Kim, H.-S., Kang, D., Kim, H., Lee, N., Yun, J., Kim, Y.-J., Lee, K.M., Kim, J.-H., Kim, H.-R., Hwang, Y.-I., Jo, C.H. & Kim, J.-H. (2021) CTRP3 Exacerbates Tendinopathy by Dysregulating Tendon Stem Cell Differentiation and Altering Extracellular Matrix Composition. Science Advances, 7, eabg6069. https://doi.org/10.1126/sciadv.abg6069.
- Zagryazhskaya-Masson, A., Monteiro, P., Macé, A.-S., Castagnino, A., Ferrari, R., Infante, E., Duperray-Susini, A., Dingli, F., Lanyi, A., Loew, D., Génot, E. & Chavrier, P. (2020) Intersection of TKS5 and FGD1/CDC42 Signaling Cascades Directs the Formation of Invadopodia. Journal of Cell Biology, 219. https://doi.org/10.1083/jcb.201910132.
- Wang, Q., Notay, K., Downey, G.P. & McCulloch, C.A. (2020) The Leucine-Rich Repeat Region of CARMIL1 Regulates IL-1-Mediated ERK Activation, MMP Expression, and Collagen Degradation. Cell Reports, 31, 107781. https://doi.org/10.1016/j.celrep.2020.107781.
- Pedersen, N.M., Wenzel, E.M., Wang, L., Antoine, S., Chavrier, P., Stenmark, H. & Raiborg, C. (2020) Protrudin-Mediated ER–Endosome Contact Sites Promote MT1-MMP Exocytosis and Cell Invasion. Journal of Cell Biology, 219. https://doi.org/10.1083/jcb.202003063.
- Kim, S.-K., Jang, S.D., Kim, H., Chung, S., Park, J.K. & Kuh, H.-J. (2020) Phenotypic Heterogeneity and Plasticity of Cancer Cell Migration in a Pancreatic Tumor Three-Dimensional Culture Model. Cancers, 12, 1305. https://doi.org/10.3390/cancers12051305.
- Lee, Y.H., Seo, E.K. & Lee, S.-T. (2019) Skullcapflavone II Inhibits Degradation of Type I Collagen by Suppressing MMP-1 Transcription in Human Skin Fibroblasts. International Journal of Molecular Sciences, 20. https://doi.org/10.3390/ijms20112734.
- Ferrari, R., Martin, G., Tagit, O., Guichard, A., Cambi, A., Voituriez, R., Vassilopoulos, S. & Chavrier, P. (2019) MT1-MMP Directs Force-Producing Proteolytic Contacts That Drive Tumor Cell Invasion. Nature Communications, 10, 4886. https://doi.org/10.1038/s41467-019-12930-y.
- Erami, Z., Heitz, S., Bresnick, A.R. & Backer, J.M. (2019) PI3Kβ Links Integrin Activation and PI(3,4)P2 Production during Invadopodial Maturation. Molecular Biology of the Cell, 30, 2367–2376. https://doi.org/10.1091/mbc.E19-03-0182.
- Bayarmagnai, B., Perrin, L., Esmaeili Pourfarhangi, K., Graña, X., Tüzel, E. & Gligorijevic, B. (2019) Invadopodia-Mediated ECM Degradation Is Enhanced in the G1 Phase of the Cell Cycle. Journal of Cell Science, 132. https://doi.org/10.1242/jcs.227116.
- Yuda, A. & McCulloch, C.A. (2018) A Screening System for Evaluating Cell Extension Formation, Collagen Compaction, and Degradation in Drug Discovery. SLAS DISCOVERY: Advancing the Science of Drug Discovery, 23, 132–143. https://doi.org/10.1177/2472555217733421.
- Castagnino, A., Castro-Castro, A., Irondelle, M., Guichard, A., Lodillinsky, C., Fuhrmann, L., Vacher, S., Agüera-González, S., Zagryazhskaya-Masson, A., Romao, M., El Kesrouani, C., Noegel, A.A., Dubois, T., Raposo, G., Bear, J.E., Clemen, C.S., Vincent-Salomon, A., Bièche, I. & Chavrier, P. (2018) Coronin 1C Promotes Triple-Negative Breast Cancer Invasiveness through Regulation of MT1-MMP Traffic and Invadopodia Function. Oncogene, 37, 6425–6441. https://doi.org/10.1038/s41388-018-0422-x.
- Mezawa, M., Pinto, V.I., Kazembe, M.P., Lee, W.S. & McCulloch, C.A. (2016) Filamin A Regulates the Organization and Remodeling of the Pericellular Collagen Matrix. The FASEB Journal, 30, 3613–3627. https://doi.org/10.1096/fj.201600354RR.
- Lodillinsky, C., Infante, E., Guichard, A., Chaligné, R., Fuhrmann, L., Cyrta, J., Irondelle, M., Lagoutte, E., Vacher, S., Bonsang-Kitzis, H., Glukhova, M., Reyal, F., Bièche, I., Vincent-Salomon, A. & Chavrier, P. (2016) P63/MT1-MMP Axis Is Required for in Situ to Invasive Transition in Basal-like Breast Cancer. Oncogene, 35, 344–357. https://doi.org/10.1038/onc.2015.87.
- Lagoutte, E., Villeneuve, C., Lafanechère, L., Wells, C.M., Jones, G.E., Chavrier, P. & Rossé, C. (2016) LIMK Regulates Tumor-Cell Invasion and Matrix Degradation Through Tyrosine Phosphorylation of MT1-MMP. Scientific Reports, 6. https://doi.org/10.1038/srep24925.
- Daubon, T., Spuul, P., Alonso, F., Fremaux, I. & Génot, E. (2016) VEGF-A Stimulates Podosome-Mediated Collagen-IV Proteolysis in Microvascular Endothelial Cells. Journal of Cell Science, 129, 2586–2598. https://doi.org/10.1242/jcs.186585.
- Marchesin, V., Castro-Castro, A., Lodillinsky, C., Castagnino, A., Cyrta, J., Bonsang-Kitzis, H., Fuhrmann, L., Irondelle, M., Infante, E., Montagnac, G., Reyal, F., Vincent-Salomon, A. & Chavrier, P. (2015) ARF6-JIP3/4 Regulate Endosomal Tubules for MT1-MMP Exocytosis in Cancer Invasion. The Journal of Cell Biology, 211, 339–358. https://doi.org/10.1083/jcb.201506002.
- Arora, P.D., Wang, Y., Bresnick, A., Janmey, P.A. & McCulloch, C.A. (2015) Flightless I Interacts with NMMIIA to Promote Cell Extension Formation, Which Enables Collagen Remodeling. Molecular Biology of the Cell, 26, 2279–2297. https://doi.org/10.1091/mbc.E14-11-1536.
- Orgaz, J.L., Pandya, P., Dalmeida, R., Karagiannis, P., Sanchez-Laorden, B., Viros, A., Albrengues, J., Nestle, F.O., Ridley, A.J., Gaggioli, C., Marais, R., Karagiannis, S.N. & Sanz-Moreno, V. (2014) Diverse Matrix Metalloproteinase Functions Regulate Cancer Amoeboid Migration. Nature Communications, 5, 1–13. https://doi.org/10.1038/ncomms5255.
- Juin, A., Di Martino, J., Leitinger, B., Henriet, E., Gary, A.-S., Paysan, L., Bomo, J., Baffet, G., Gauthier-Rouvière, C., Rosenbaum, J., Moreau, V. & Saltel, F. (2014) Discoidin Domain Receptor 1 Controls Linear Invadosome Formation via a Cdc42–Tuba Pathway. Journal of Cell Biology, 207, 517–533. https://doi.org/10.1083/jcb.201404079.
- Haeger, A., Krause, M., Wolf, K. & Friedl, P. (2014) Cell Jamming: Collective Invasion of Mesenchymal Tumor Cells Imposed by Tissue Confinement. Biochimica Et Biophysica Acta, 1840, 2386–2395. https://doi.org/10.1016/j.bbagen.2014.03.020.
- Gligorijevic, B., Bergman, A. & Condeelis, J. (2014) Multiparametric Classification Links Tumor Microenvironments with Tumor Cell Phenotype. PLoS biology, 12, e1001995. https://doi.org/10.1371/journal.pbio.1001995.
- Wolf, K., te Lindert, M., Krause, M., Alexander, S., te Riet, J., Willis, A.L., Hoffman, R.M., Figdor, C.G., Weiss, S.J. & Friedl, P. (2013) Physical Limits of Cell Migration: Control by ECM Space and Nuclear Deformation and Tuning by Proteolysis and Traction Force. The Journal of Cell Biology, 201, 1069–1084. https://doi.org/10.1083/jcb.201210152.
- Monteiro, P., Rossé, C., Castro-Castro, A., Irondelle, M., Lagoutte, E., Paul-Gilloteaux, P., Desnos, C., Formstecher, E., Darchen, F., Perrais, D., Gautreau, A., Hertzog, M. & Chavrier, P. (2013) Endosomal WASH and Exocyst Complexes Control Exocytosis of MT1-MMP at Invadopodia. The Journal of Cell Biology, 203, 1063–1079. https://doi.org/10.1083/jcb.201306162.
Include figures
1
Include data sheet
1
Place your order or ask questions about this product.





